Section: New Results
A model of genome size evolution
Participants: G. Beslon, C. Knibbe, S. Fisher
Even though numerous genomic sequences are now available, evolutionary mechanisms that determine genome size, notably their fraction of non-coding DNA, are still debated. In particular, although several mechanisms responsible for genome growth (proliferation of transposable elements, gene duplication and divergence, etc.) were clearly identified, mechanisms limiting the overall genome size remain unclear.
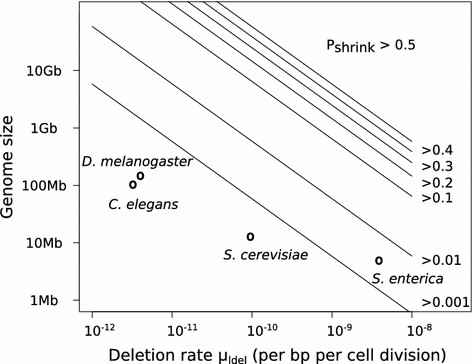
In collaboration with Samuel Bernard (Inria Dracula Team and Institut Camille Jordan, UMR CNRS 5208, Lyon), we have developed a model for genome size evolution that takes into account both local mutations such as small insertions and small deletions, and large chromosomal rearrangements such as duplications and large deletions. We introduced the possibility of undergoing several mutations within one generation. The model, albeit minimalist, revealed a non-trivial spontaneous dynamics of genome size: in the absence of selection, an arbitrary large part of genomes remains beneath a finite size, even for a duplication rate 2.6-fold higher than the rate of large deletions, and even if there is also a systematic bias toward small insertions compared to small deletions. Specifically, we showed that the condition of existence of an asymptotic stationary distribution for genome size non-trivially depends on the rates and mean sizes of the different mutation types. We also gave upper bounds for the median and other quantiles of the genome size distribution, and argue that these bounds cannot be overcome by selection. Taken together, these results show that the spontaneous dynamics of genome size naturally prevents it from growing infinitely, even in cases where intuition would suggest an infinite growth. This work was part of Stephan Fischer's PhD thesis, which was defended in December 2013.
|
This year, using quantitative numerical examples with parameters taken from biological data, we showed that, in practice, a shrinkage bias appears very quickly in genomes undergoing mutation accumulation, even though DNA gains and losses appear to be perfectly symmetrical at first sight. This spontaneous dynamics provides the genome with a stability-related size limit below which it can be influenced by other evolutionary forces (selection, drift, biases, ...).
All this work has been published this year [15] , and is already mentioned as "most read article" by Springer.